News
-
- 11.28, 2024
- Dr. Kazuki Saito, Group Director and Dr. Takayuki Tohge, Senior Visiting Scientist has been selected for Highly Cited Researchers2024
- Highly Cited Researchers 2024
-
- 11.30, 2023
- Dr. Kazuki Saito, Group Director and Dr. Takayuki Tohge, Visiting Scientist has been selected for Highly Cited Researchers2023
- Highly Cited Researchers 2023
- 10.3, 2023
- Dr. Amit Rai, Researcher was featured in "RIKEN People".
-
- 12.8, 2022
- Dr. Kazuki Saito, Group Director, received "Shimadzu Award 2022"
Research Outline
Metabolomics and Phytochemical Genomics
Metabolomics involves in the identification and quantification of all metabolites in a cell, and correlating these to genomic functions. In this research group, we perform high-throughput metabolite analyses, and develop technologies for the comprehensive analysis of metabolites, mainly using high-resolution mass spectrometry. We also collaborate with other research teams within and outside the Center on non-targeted analyses and the identification of unknown gene functions. The various compounds produced by plants are important for the existence of the plant itself, and also play a vital role in our lives. We are investigating the basic principles behind the wide variety of plant production functions, using Arabidopsis as a model. We are also analyzing the production systems for specific products in crops, medicinal plants and other useful plants at a genome level. Another important aspect of our research is the direct application of the basic findings from these results to biotechnological application of plants.
Contents of Research
- Development of analytical techniques for mass spectrometry.
- Database construction and bioinformatics for metabolomic data analysis.
- Metabolomic analysis for plant biology through collaboration with other groups.
- Comprehensive identification of genes involved in primary and secondary metabolite biosynthesis using post-genomic methods in Arabidopsis
- Identification of new genes and networks involved in the biosynthesis and accumulation of flavonoids, alkaloids, terpenoids, sulfur-containing constituents, amino acids and other substances in non-model plants, and their application to the production of useful substance
Results of Research
- The metabolomics pipeline was constructed for the comprehensive analysis of various plant metabolites including amino acids, organic acids, nucleotides, complex lipid and secondary metabolites. The pipeline consists of GC (gas chromatography)-, CE (capillary electrophoresis)-, and LC (liquid chromatography)-MS branches with a data processing techniques based on the multivariate analysis.
- Large number of chromatogram and mass spectrum of plant metabolites were collected for an information basis of the plant metabolome analysis. The data were open to the public via MassBank, a mass spectrum database (JST/BIRD).
- Behaviors of the plant primary metabolism were compared among the wild-type and metabolic mutants of Arabidopsis by the metabolite-metabolites correlation network analysis. The data mining with a combination of transcriptome data revealed the candidate genes responsible for the regulation of primary metabolism in plants.
- We have established a database 'AtMetExpress development', which accumulate the metabolome data of developmentally-different Arabidopsis tissues.
- The function of flavanoid and anthocyanin biosynthesis related genes of Arabidopsis were identified by the integration analysis of metabolomics, transcriptomics and biochemical approaches.
- Metabolomic analysis as collaborative projects with internal and external groups for a variety of plant functions with Arabidopsis thaliana and crops.

Fig.1 Mass spectrometers using for metabolomic analysis
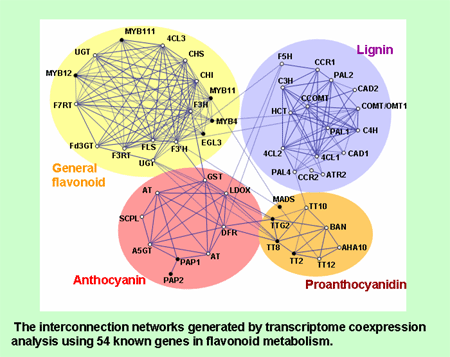
Fig.2 Co-expression gene network of plant secondary metabolism








